| |
FOXJ1
forkhead box J1| Gene | | Official Symbol | FOXJ1 | | Official Full Name | forkhead box J1 | | CGNC ID | 14612 | | also known as | forkhead box protein J1 | | gene type | protein-coding |
| | Genomic Map | | | Sequence Information | | | Gene Expression | | In Situ Hybridization | GEISHA | | EST Profile | |
| | Orthology | | Entrez Gene | Ensembl Gene | Genetic Phenotypes | MOD | | Fruit Fly | | | | | | Human | 2302 | ENSG00000129654 | MIM:607154 | MIM:602291 | | Mouse | 15223 | ENSMUSG00000034227 | All phenotypic alleles (4):Targeted, knock-out(1) Targeted, other(3) | MGI:1347474 | | Xenopus | 493505, 734957, 496834, 399038, 399039 | | | 919737, 853647 | | Zebrafish | 767737, 494105 | ENSDARG00000059545, ENSDARG00000088290 | ZDB-GENE-060929-1178, ZDB-GENE-041212-76 | ZFIN:ZDB-GENE-060929-1178, ZFIN:ZDB-GENE-041212-76 |
| | Gene Ontology | | Molecular Function | | | Biological Process | | | Cellular Component | |
| | Links to other databases | |
Entry 1| GEISHA Id | FOXJ1.UApcr | Data Source | GEISHA ISH Analysis | | Forward cDNA Template Primer | 5’-TGGCAATGAGTACCTGTCCG-3’ | | Reverse cDNA Template Primer | 5’-ATTAACCCTCACTAAAGGGAACGGTGCTGACATTGACG-3’ | cDNA Template Length | 886 | | Complete cDNA Template Probe | show TGGCAATGAGTACCTGTCCGGCTCTGTCAGCATGGAGCAGCTGTTTGAGCTCGGTGACACCTCGCTGCCCGCAGACGTC
AACGAATGGAGCGCTGTGGGATCCTTTCTATAAGCAAAGGACCGAACCAAACCCATGGGGGCTTTGGGAGGACGCTCCC
TTTGTGCTGCTGGAACTTGCAATGGACAATATTTTCTACGGACAGACTCCCCCAGCAGCTTGATCAGCTTCATGGTAGG
GTTTATTCATAGACACACCAATGGGGAGAACACAGTAACACAAAAAATTGCTTGAAATATCTTCAAAGGACACACCAGA
AGCCCGAGGTCCACCGCTTTCCTCAAGAGAAGAGGTAGAACAGTACTTATTTTTTTACTTTTTCAGGAACAAAATGTAC
ATTCCCACTTTTCCTTGCATGGAAATGGACACGGCATACAGTCCTCCTCTGCAGACCCAACAGACAAGATTCCTGGTGG
CTTCGGACTCCATAGACGACGTGCAATCTTCTCACTCCTTCTTCTCTCTGTTCTTCTCTCTCTTTCTGTCCGGGTTCGT
TTGTAGGTGTAGCTAATTAGTTAGCAGGCAAACAGAGTCCCCTCTGGATTCTCCAAGTTCAACCTCCCTCCTGACTTGT
GCCTGGGGAAGAAGGAAAGATCCGTGCATTCAGTCTGCCTCTTTAGATGTTTTTATATAATCTATTATATATTATATAT
TCAAAGAAACCTATATTTTTAGCATTAGGACCTCTTTTCAGAGTGGGGTAGGGGAAATGTAGGTTGGGGTTGTTTTGTA
CTTGGGCTTATGTTAGTCGTTCTGCAAACCAAGACCTGCACAGTGGCTTTGGTCAGCATTGGGTCACCCAACTCCACGT
CAATGTCAGCACCGTTC |
Entry 2| GEISHA Id | FOXJ1.Cruz.2010 | Data Source | Publication | | Complete cDNA Template Probe | show ATGGCTGAGGGCTGGCTGAGCCTGCGGGCGGCCGAGGGGCAGCAGGGCGGCATGGGCGACTCGAGCAACCTGGACGACA
GCCTGACCAGCCTGCAGTGGCTGCAGGAGTTCTCCATCATCAACGCCAACGTGGGCAAATCCTCCTCGTGCCCCAGCGG
CCCCGATCCTCACGATTGCCACCACGTCCCCGGCTTCGCCGCTCCGTGCTCGCCCCTGGCGGCCGACCCGGCGTGTATG
GGGATGCCCCACACCCCGGGCAAACCCACCTCCTCCTCCACCTCGAGGACGGCACACCCGGCTTTGCAGGCTCAGCCGC
CCGAGGACATCGACTACAAGACCAACCCGCACGTGAAGCCGCCCTACTCCTACGCCACTCTCATCTGCATGGCGATGGA
AGCCAGCAAGAAGACCAAAATCACCCTGTCCGCCATCTACAAGTGGATTACCGACAACTTCTGCTACTTCCGACACGCC
GACCCCACCTGGCAGAACTCCATCCGACACAACCTGTCCTTGAACAAGTGCTTCATCAAAGTGCCCCGCGAGAAGGACG
AGCCGGGGAAAGGCGGCTTTTGGAAGATCGACCCTCAGTACGCCGACCGGCTGATGAACGGGGCCTTCAAGAAGCGGCG
GATGCCCCCGGTGCAGATCCACCCGGCCTTCACCAACAGGGCCCAGCACCAAGCGTGCGGCGTCGGCGGGCCGCTGCCT
TCCAGCTGCGCCAACAGCAACGTCCTCAACGTCAACACGGAGTCCCAGCAGCTGCTGAAGGAGTTTGAAGAGGTCACCG
GAGAGCAGAGTTGGGATGGGAAGAGCGGCCGCAAGCGCAAGCAGCCCTTACCCAAACGCACGGCCAAGGCGGCCCGGCT
GCTCAACACGGCCCTGCTCACCCAGGAGGAGCAGACCGAGCTGGGCTCGCTGAAGGGCAACTTCGACTGGGAGGTTGTC
TTTGACACCAACCTCAATGGAGATTTCACCACGTTTGAGGAGCTGGAGCTCACGCCGCCCATCAGCCCCATTGGGCGTG
ATGTTGACCTGACGGGCCACGGATACCACGACGGTGCACAGGAGTGGTGCGTGGCCGGGCCCGAGCAGCCCCTCACCGA
GCCCAACCACAACAACCTGGACTTTGACGAGACCTTCATGGCCACCTCCTTCCTGCAGCACCCCTGGGATGACGATGGC
AATGAGTACCTGTCCGGCTCTGTCAGCATGGAGCAGCTGTTTGAGCTCGGTGACACCTCGCTGCCCGCAGACGTCAACG
AATGGAGCGCTGTGGGATCCTTTCTATAA | | Citation | Cruz C, Ribes V, Kutejova E, Cayuso J, Lawson V, Norris D, Stevens J, Davey M, Blight K, Bangs F, Mynett A, Hirst E, Chung R, Balaskas N, Brody SL, Marti E, Briscoe J. Foxj1 regulates floor plate cilia architecture and modifies the response of cells to sonic hedgehog signalling. Development. 2010 Dec;137(24):4271-82. | | Copyright | Copyright retained by the authors. Permission to reprint the images has been requested. | | Comments | Insufficient information provided in publication to verify exact sequence used to synthesize probe. Sequence below was obtained from NCBI (acc # XM_001233326). |
| Stage | Image (click image to view full size) | Location (click to highlight) | Comments (click to highlight) | | 19-22 | 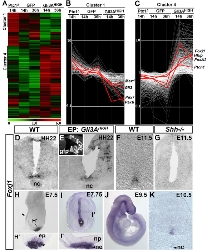 | | stage 22 |
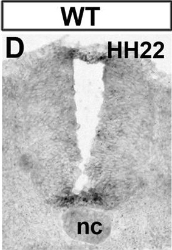 | | stage 22 |
| Neural Plate/Tube
|
MouseFrogFruit FlyZebra FinchZebrafish |
|