| |
PSMA7
proteasome subunit alpha 7| Gene | | Official Symbol | PSMA7 | | Official Full Name | proteasome subunit alpha 7 | | CGNC ID | 49262 | | also known as | proteasome subunit alpha type-7|GPRO-28|proteasome (prosome, macropain) subunit, alpha type, 7|proteasome 28 kDa subunit homolog | | gene type | protein-coding |
| | Genomic Map | | | Sequence Information | | | Gene Expression | | | Orthology | | | Gene Ontology | | | Links to other databases | |
| GEISHA Id | PSMA7.Olivera-Martinez.2014 | Data Source | Publication | | Complete cDNA Template Probe | show GGCGGCGCGCGGCGCCAGCCATGAGCTACGACCGGGCCATCACCGTCTTCTCTCCGGACGGGCACCTCTTCCAGGTGGA
GTACGCCCAGGAGGCGGTGAAGAAGGGCTCTACGGCGGTTGGGGTCAGAGGGAAGGACATCGTTGTTCTCGGCGTGGAG
AAGAAGTCTGTGGCAAAACTTCAGGATGAAAGAACTGTGCGGAAGATCTGCGCCCTCGATGACAATGTCTGCATGGCTT
TTGCAGGGCTCACAGCTGATGCCAGAATAGTTATAAACAGAGCTCGTGTAGAATGTCAGAGCCACAGACTCACCGTGGA
GGATCCGGTCACCGTGGAGTACATCACGCGTTACATCGCCAGCCTGAAGCAGAGGTATACCCAAAGCAAACGTCGCAGA
CCTTTTGGTATCTCTGCTCTGATTGTGGGATTTGACTTTGATGGAACCCCCCGGCTGTACCAGACTGACCCCTCTGGCA
CATACCATGCTTGGAAGGCTAATGCCATTGGCAGAGGAGCTAAATCTGTACGTGTGGAATTCGAGAAAAACTATACTGA
TGAAGCCATTGAAACAGATGATCTGACCATTAAGCTTGTCATCAAGGCTCTTCTTGAGGTTGTGCAGTCTGGTGGGAAG
AACATCGAGCTGGCTGTTATGAGAAGAGATCAGCCCCTGAAGATTCTAACGTCCCCTGAAGAAATTGAGAAGTATGTTG
CTGAAATTGAAAAAGAAAAGGAAGAAAATGAAAAGAAAAAGCAGAAGAAGACATCATGATGAACCAAATCCAGCACCTG
CTGAGCTGCTTCTTTTTAACTGAAGCAATGGGTTATTCTGTATAGACAAGATGTAGGCATTTCCGTTTTATCATACTGT
GCCCTCTGGAGACCTACAATAAACCTACTGATTTTTAGCCTTAAAAAAACGAGTTTTTTTTTTTTTTTTTTTTTTTTTT
TTTTTTT | | Citation | Isabel Olivera-Martinez, Nick Schurch, Roman A Li, Junfang Song, Pamela A Halley, Raman M Das, Dave W Burt, Geoffrey J Barton, Kate G Storey. Major transcriptome re-organisation and abrupt changes in signalling, cell cycle and chromatin regulation at neural differentiation in vivo. Development 2014 Aug;141(16):3266-76. Epub 2014 Jul 25. DOI: 10.1242/dev.112623. | | Copyright | Copyright © 2014. Published by The Company of Biologists Ltd. | | Comments | As insufficient information is provided in publication to verify exact sequence used to synthesize probe, the sequence above was obtained from NCBI (accession number NM_204613.1 ). |
| Stage | Image (click image to view full size) | Location (click to highlight) | Comments (click to highlight) | | 7-10 |
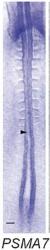 | | stage 10 |
| Neural Plate/Tube
| | all | | |
| | Data from NCBI Unigene EST Profile
Unigene ID: Gga.2045 |
MouseFrogFruit FlyZebra FinchZebrafish |
|